
Nitu Kumar
University of the West Indies, Trinidad and Tobago
Title: Isolation and molecular characterization of Salmonella enterica strains from poultry in Trinidad
Biography
Biography: Nitu Kumar
Abstract
Statement of the Problem: Salmonella is one of the leading causes of foodborne illnesses worldwide impacting on public health in Trinidad. Salmonella infections are usually associated with the consumption of contaminated food products and infections in humans generally lead to acute gastroenteritis that may become complicated depending on the strain, serotype and host-specific factors. Changes in agricultural practices and antimicrobial misuse in food producing animals may be accelerating factors for the evolution of more virulent and multidrug-resistant strains. The objective of this study was to identify types of antimicrobial resistance and their associated genes, virulence-associated genes, and to analyze the pulsed-field gel electrophoresis (PFGE) patterns of Salmonella enterica serotypes isolated from poultry sale outlets in Trinidad.
Methodology: A total of 1503 caecal samples were collected from different ‘pluck shops’ in Trinidad and confirmed to be Salmonella using standard techniques. PCR-based assays were performed on 88 Salmonella isolates to detect 13 virulence-associated genes. Isolates were further assessed for their susceptibility to five antimicrobial agents using tube dilution method to determine the minimum inhibitory concentration (MIC). Resistant isolates were subsequently examined for the presence of 11 resistance genes. PFGE was used, after DNA digestion by XbaI, to investigate the genetic relatedness among Salmonella enterica isolates recovered from poultry in Trinidad.
Findings: 11 (84.6%) of the 13 virulence genes investigated were detected and their frequency ranged from 1.3% (sefC) to 84.2% (mgtB). Only 4 (36.4%) of the 11 resistance genes tested for were detected and their frequency ranged from 1.3% (ampicillin) to 63.2% (quinolones). Of the five antimicrobial agents tested for MIC, the range was 7.5 µg/ml (gentamicin), 10-20 µg/ml (streptomycin), 1.25-10 µg/ml (ceftriaxone), 20-80 µg/ml (kanamycin) and 20-40 µg/ml (ampicillin). The PFGE patterns of Salmonella spp. of the same serotypes from chickens and ducks sampled from various outlets were distinctly different.
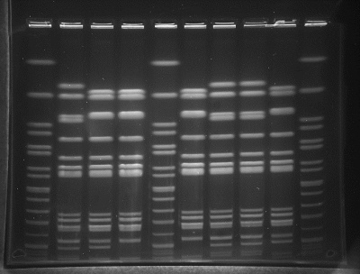
Pulsed Field gel electrophoresis (PFGE) patterns of Salmonella enterica isolates following digestion by restriction enzyme XbaI
Lanes 1: 5 10: Standard controls (Salmonella serotype Braenderup H9812 )
Lane 2: SD21 Salmonella Sub Species Enterica I from outlet S
Lane 3: A149 Salmonella Sub Species Enterica I from outlet A
Lane 4: A159 Salmonella Sub Species Enterica I from outlet A
Lane 6: R41 Salmonella Sub Species Enterica I from outlet R
Lane 7: SD22 Salmonella Typhimurium from outlet S
Lane 8: SD55 Salmonella Typhimurium from outlet S
Lane 9: A142 Salmonella Typhimurium from outlet A

